Structural Virology
Understanding virus structure is crucial in order to decipher important features of the viral lifecycle as well as to open up new avenues to interfere with virus assembly.
Structural conservation of retrovirus capsid assemblies
Specifically, we are interested in the conservation and diversity of retroviral capsid assemblies. Structural approaches have been suggested to be more optimally suited than sequence alignment methods to define evolutionary relationships, given the stronger constraints in structure space (i.e. the lower number of possibilities of protein folds that can assemble meaningful structures)(see Abrescia et al., 2012). This is highlighted when looking at retroviral CA structures where despite a low primary sequence conservation, the tertiary structure of CA proteins is remarkably similar.
The retrovirus family includes important human pathogens, such as HIV-1, but also other important retroviral model systems. Our recent advances in cryo-EM and cryo-ET methodology and the ongoing improvement of retroviral in vitro assembly systems have now made it possible to provide unprecedented high-resolution insights into retroviral Gag and CA structures (see Publications). We continue to study how different retroviruses asssemble immature and mature virus particles and to what extent their assembly and maturation mechanism differ.
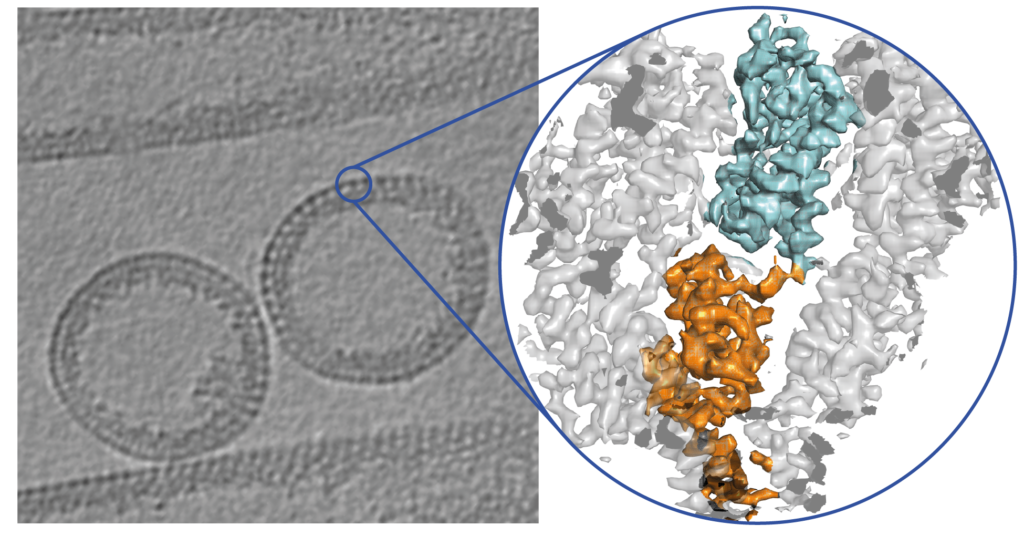
Structural biology of poxviruses

Poxviruses are among the largest double-stranded DNA viruses with members such as variola virus, monkeypox virus and the vaccination strain Vaccinia virus (VACV). The poxvirus core is one of the uniting factors in all the infectious poxvirus forms and fulfils key roles in the virus lifecycle, such as the protection of the viral genome. Knowledge about the structural proteins that form the viral core has remained sparse, impeded by the molecular complexity of poxviruses, such as the large genome with more than 200 proteins, the large virus size and its variable dimensions.
Poxviruses represent a challenging and rewarding target for our cryo-EM approaches to answer open questions on the core architecture. Using a multi-modal cryo-electron microscopy (cryo-EM) approach our lab could elucidate the structural determinants of the VACV core (Datler, Hansen et al., 2024).
Due to the dependence of poxviruses on the actin cytoskeleton for viral spread, they also are a valuable model system to study actin-related processes (see for example Fäßler et al., 2023).